Capture-SELEX
In classic magnetic beads-based SELEX (MB-SELEX) methods, the target is immobilized on the substrate (generally magnetic beads) and then the free nucleic acid library is added for specific binding. The bound oligonucleotides are separated from the target and amplified to continue in the next round of selection. However, this does not apply to a number of small-molecule targets, which are difficult to immobilize. The immobilization of small-molecule targets has limitations: (1) There are few functional groups on some small molecules that can be used for coupling, and actually there is no or lack of suitable groups that can be immobilized on the substrate for such targets. (2) During screening of immobilized targets, the native structure of the target may be compromised due to some extreme denaturing conditions such as pH and temperature. (3) After binding to the carrier matrix, it will be affected by the masking effect, and fewer binding sites are exposed, resulting in the loss of some nucleic acid sequences that can bind to the masked chemical groups. Therefore, aptamer screening using immobilized targets may reduce the success rate and efficiency inherently. Whereas the small-molecule target is difficult to immobilize, the nucleic acid library can be immobilized on the carrier, thereby developing the scheme of Capture-SELEX in 2005. The Capture-SELEX strategy addresses the limitations of some small-molecule targets that are difficult to immobilize and has been success-fully used to screen aptamers against small molecules.
Capture-SELEX is applicable to target molecules like small molecules, irons, molecules without functional groups for conjugation, which cannot be bound and immobilized to the magnetic bead.
Fusion BioLabs further develop the main process of DNA Capture-SELEX and RNA capture-SELEX screening methods to efficiently screen aptamers against small to large molecular targets. Following is the Capture-SELEX process.
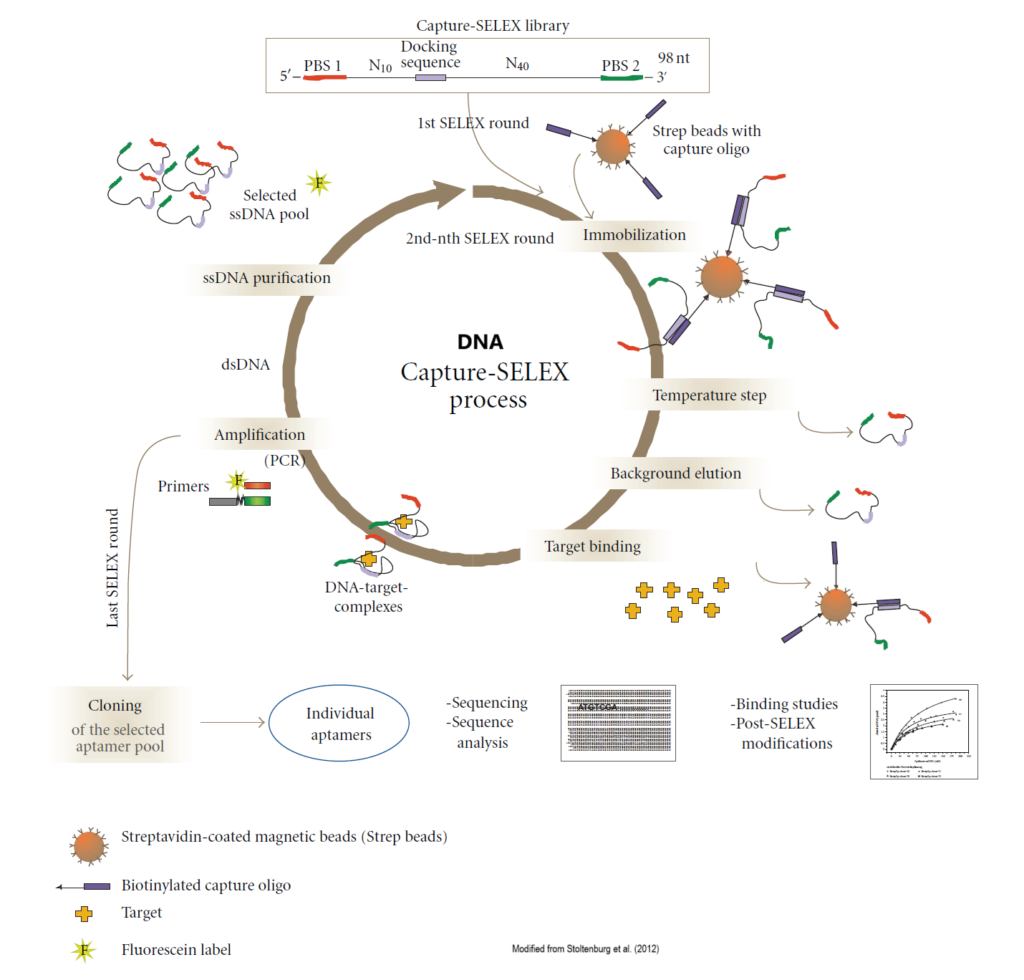
DNA Capture-SELEX
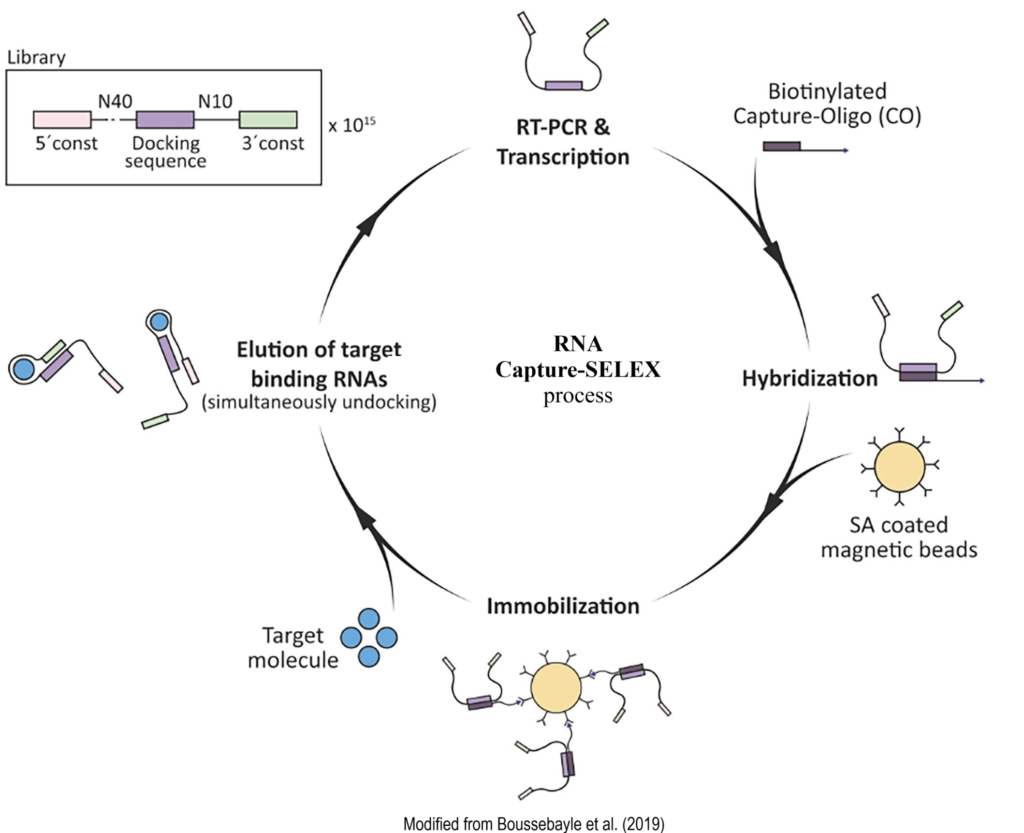
RNA Capture-SELEX
Main Steps for Capture-SELEX
- Couple the nucleic acid library to a solid-phase substrate (generally magnetic beads) via bridging fragments, which are usually located in the middle, and sometimes coincide with primer sequences of the library oligo.
- Partition of target-binding and nonbinding sequences: Co-incubation of the target with the immobilized nucleic acid pool, target-induced conformational switching results in the detachment of bound sequences from the surface of the solid substrate, while weakly or unbound sequences remain on it.
- Enrichment: the sequences that specifically recognize the target are enriched and collected, and the enrichment library was amplified and repeatedly selected until the desired high-affinity aptamer was obtained.
Aptamer Development through Capture-SELEX
Phase I: Library Enrichment via Capture-SELEX
Iterative rounds (10-15 rounds) of screening an initial random library against a target for positive selection, and negative and/or counter-selection screening against sample matrix or counter-target(s) for specificity.
Criteria: Enrichment 8-30%.
Deliverables: full enrichment progress report
Timeline: 2-3 weeks
Phase II: Sanger Sequencing & Bioinformatic Analysis
Last round of positive binding candidates cloning into sequencing vector for polyclonal preparation and picking up to 48 clones (Basic Package and Standard Package) for Sanger sequencing.
Deliverables: Aptamer candidate sequence information, sequence alignment, family cohort, secondary structure analysis, docking simulation et al..
Timeline: 1-2 weeks
Phase III: NGS Sequencing & Bioinformatic Analysis (optional)
Next-Generation Sequencing (NGS) of last positive round and representative rounds, negative binding rounds, and control of initial non-enriched library. Bioinformatically analyzing to identify the potential top aptamer candidates by ranking the aptamers based on their enrichment ratio or fold-change between positive and control/negative rounds.
Deliverables: NGS analysis full report
Timeline: 2-3 weeks
Phase IV: Aptamer Candidate Synthesis, Affinity Determination and Specificity Assessment
Depending on the results of data analysis, up to 96 aptamer hits (Premium Package or Premium Plus Package) showing high enrichment profiles and different structural families will be synthesized in small-scale to determine top candidates for further characterization. Top aptamer candidates will have a high enrichment ratio and high statistical significance in the positive rounds compared to control.
Final ranking and validation to confirm their binding affinity and specificity: each top aptamer candidate (up to 8) will be synthesized and quantitatively tested for KD affinities via GO-SELX, Capture-SELEX, AuNP-SELEX, biolayer interferometry (BLITz), surface plasmon resonance (SPR), enzyme-linked aptamer-sorbent assay (ELASA), or gel shift assays.
Deliverables: 10 nmol each aptamer candidates, and full assay report showing KD value and specificity for each aptamer candidates.
Timeline: 3-4 weeks
Phase V: Aptamer Optimization & Assay Development
Depends on the criteria set by clients, the cost and timeline will be different.
Deliverables: Full assay development report and 10m nmol optimized aptamers.
Timeline: 2-4 weeks
Fusion Biolabs offer the following packages
| Basic Package | Standard Package | Premium Package | Premium plus Package | |
|---|---|---|---|---|
| Phase I: Library Enrichment via Capture-SELEX | ||||
| Phase II: Sanger Sequencing & Bioinformatic Analysis | ||||
| Phase III: NGS Sequencing & Bioinformatic Analysis | ||||
| Phase IV: Aptamer Candidate Synthesis, Affinity Determination and Specificity Assessment | ||||
| Phase V: Aptamer Optimization & Assay Development | ||||
| Package Cost* | inquiry | inquiry | inquiry | inquiry |
1) Fusion BioLabs guarantees the services;
2) Fusion BioLabs owns its proprietary processes of aptamer development. However, the client owns full rights to the aptamers Fusion BioLabs developed for the client;
3) For multiple targets aptamer development, please contact us for special pricing.
References
1. Stoltenburg R, Nikolaus N, Strehlitz B (2012) Capture-SELEX: Selection of DNA Aptamers for Aminoglycoside Antibiotics. J. Anal Methods Chem 415607.
2. Boussebayle A, Groher F, Suess B (2019) RNA-based Capture-SELEX for the selection of small molecule-binding aptamers. Methods 161: 10-15.
